Research Interests
Pif1 family helicases:
Pif1 helicases belong to an evolutionarily conserved enzyme family found from bacteria to humans. They have been best studied in the budding yeast Saccharomyces cerevisiae, but their functions have also been investigated in the fission yeast Schizosaccharomyces pombe, trypanosomes, mice, and humans. In these organisms, the Pif1 helicases are multifunctional, usually working to ensure the integrity of both the nuclear and mitochondrial genomes via their various functions in Okazaki fragment maturation, double-strand break (DSB) repair, replication fork barrier maintenance, and maintaining G-quadruplex (G4) DNA stability.
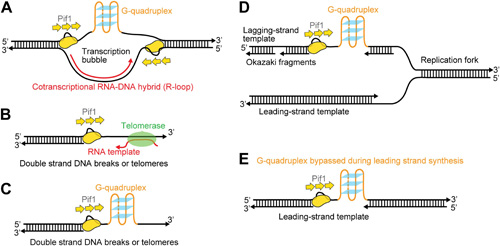
Possible cellular sites for the patrolling activity of a Pif1 monomer.
Periodic patrolling activity may keep Pif1 at its sites of in vivo action in resolving biologically relevant �R-loops� (A), displacing telomerase from 3' ssDNA ends at telomeres or DSBs (B), unwinding G4 structures on the 3' ssDNA tails at telomeres or DSBs (C), and unwinding G4 structures on the lagging and/or leading strands during DNA replication (D and E). Adapted from DOI: http://dx.doi.org/10.7554/eLife.02190.022.
Currently, we�re working on a variety of Pif1 projects to: 1) biochemically compare Pif1 activities across species, 2) determine the role(s) of a bacteriophage Pif1, and 3) utilize Pif1 helicases from thermophilic bacteria as novel reagents for both molecular biology and structural studies.
RecQ family helicases:
RecQ family helicases are conserved from bacteria to humans, with more complex organisms generally encoding multiple family members (e.g., one in E. coli and five in humans). These enzymes are well known for their roles in DNA recombination, but they also function in DNA replication, repair, telomere maintenance, and transcription, as well as have mitochondrial functions. Notably, mutations in three of the five human RecQs (BLM, WRN, and RECQ4) are associated with diseases characterized by predispositions to cancer and/or premature aging.
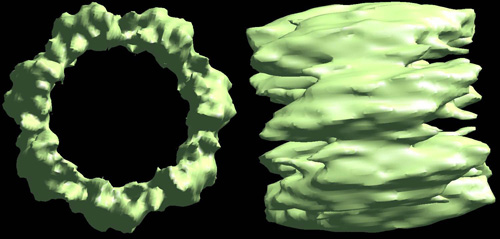
Low resolution 3D reconstruction of the S. cerevisiae Hrq1 heptameric structure.
Single particles of purified Hrq1 were semi-automatically picked from negatively stained transmission electron micrographs, rotationally aligned and averaged into class averages, and built into a 3D structure using EMAN2 software.
Our current focus in the lab is on RECQ4 and using its S. cerevisiae homolog Hrq1 as a simple model to determine why mutation of human RECQ4 leads to three different diseases: Rothmund-Thomson Syndrome, Baller-Gerold Syndrome, and RAPADILINO. We�re using biochemistry, mass spectrometry, and yeast genetics to determine the substrate specificities of these enzymes and their in vivo interactomes.
Alcohol and fermentation:
S. cerevisiae isn�t only a workhorse model organism in biology, it�s also responsible for nearly all of the industrial ethanol fermentation that takes place to give us beer, wine, the precursors to distilled spirits, and biofuel. However, other yeast species are also ethanol tolerant, including the lager yeast S. uvarum (a hybrid between S. cerevisiae and S. eubayanus) and many wild yeasts.
In collaboration with Indiana craft brewers Rob Caputo and Justin Miller, we are bioprospecting for wild yeasts with desirable brewing characteristics, brewing test batches of beer, and conducting sensory analyses of the final products. This work was originally funded by the Johnson Center for Entrepreneurship in Biotechnology and has in part resulted in the creation of a start-up company called Wild Pitch Yeast
To date, we have worked with Cardinal Spirits, Central State Brewing, the Flat 12 Bierwerks, Function Brewing, Tin Man Brewing Company, and Upland Brewing Company.
